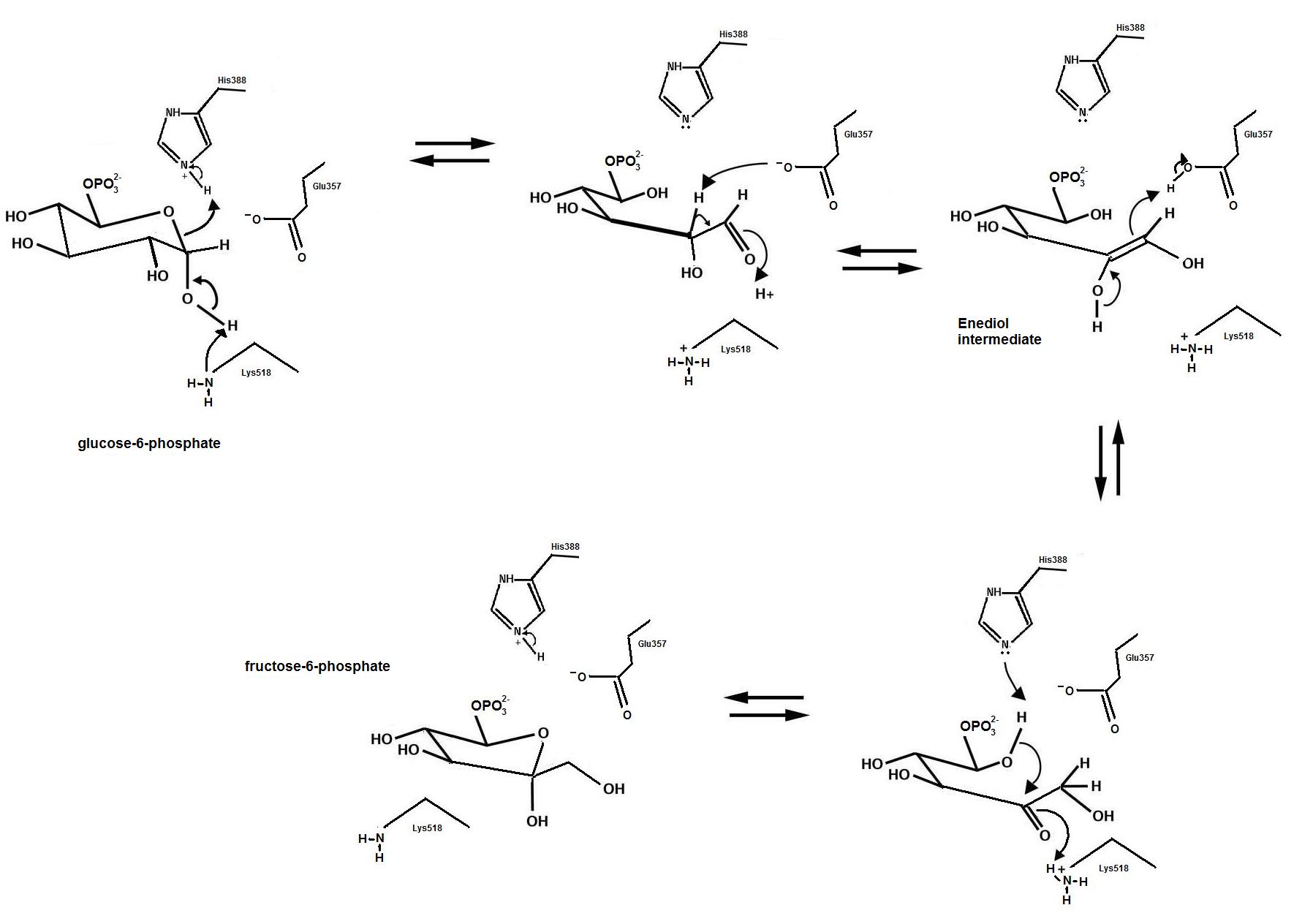
Catalytic reaction mechanism of Pseudomonas stutzeri l‐rhamnose isomerase deduced from X‐ray structures - Yoshida - 2010 - The FEBS Journal - Wiley Online Library
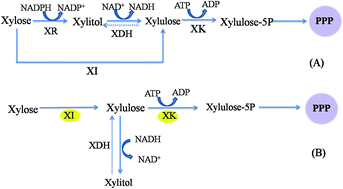
The role of a xylose isomerase pathway in the conversion of xylose to lipid in Mucor circinelloides - RSC Advances (RSC Publishing)

Locating active-site hydrogen atoms in d-xylose isomerase: Time-of-flight neutron diffraction | PNAS

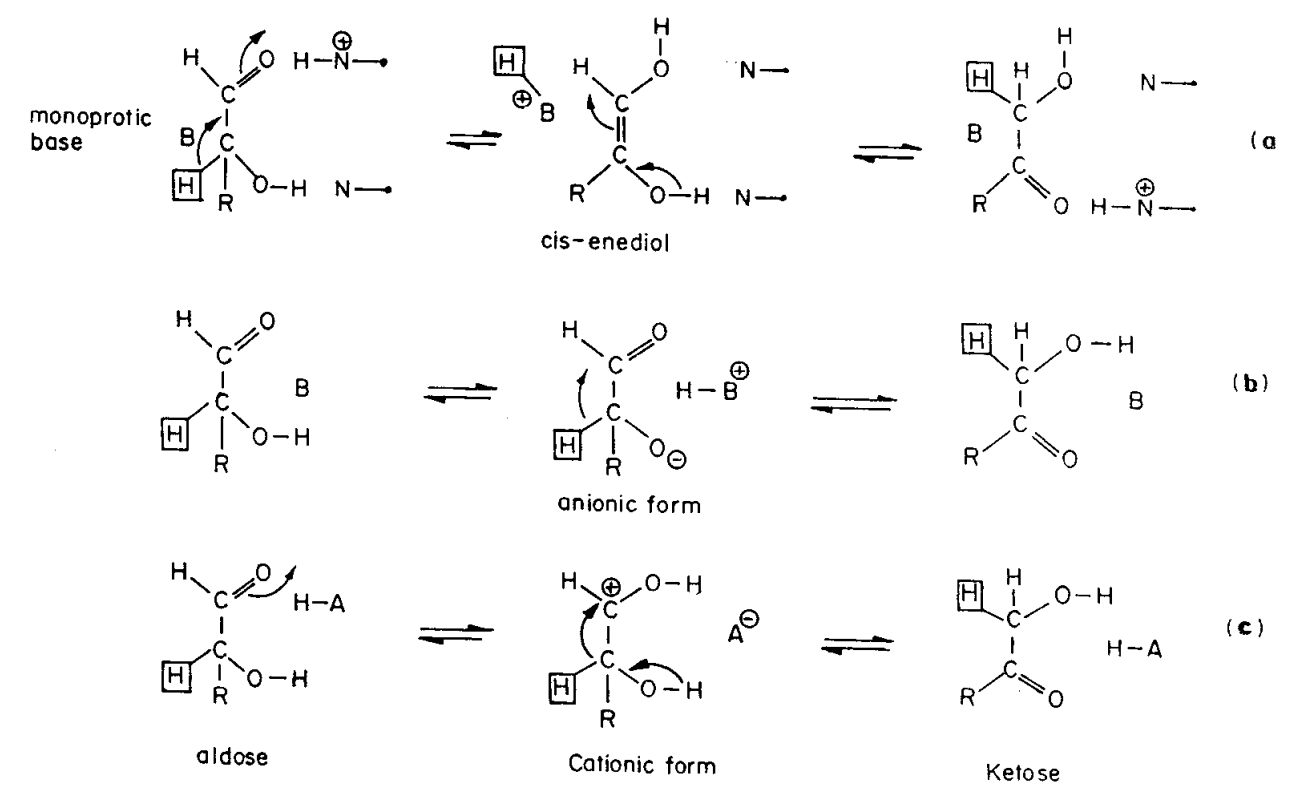
Figure 1 from Observations of reaction intermediates and the mechanism of aldose-ketose interconversion by D-xylose isomerase. | Semantic Scholar

Magnesium in PDB 2xis: A Metal-Mediated Hydride Shift Mechanism For Xylose Isomerase Based on the 1.6 Angstroms Streptomyces Rubiginosus Structures with Xylitol and D-Xylose
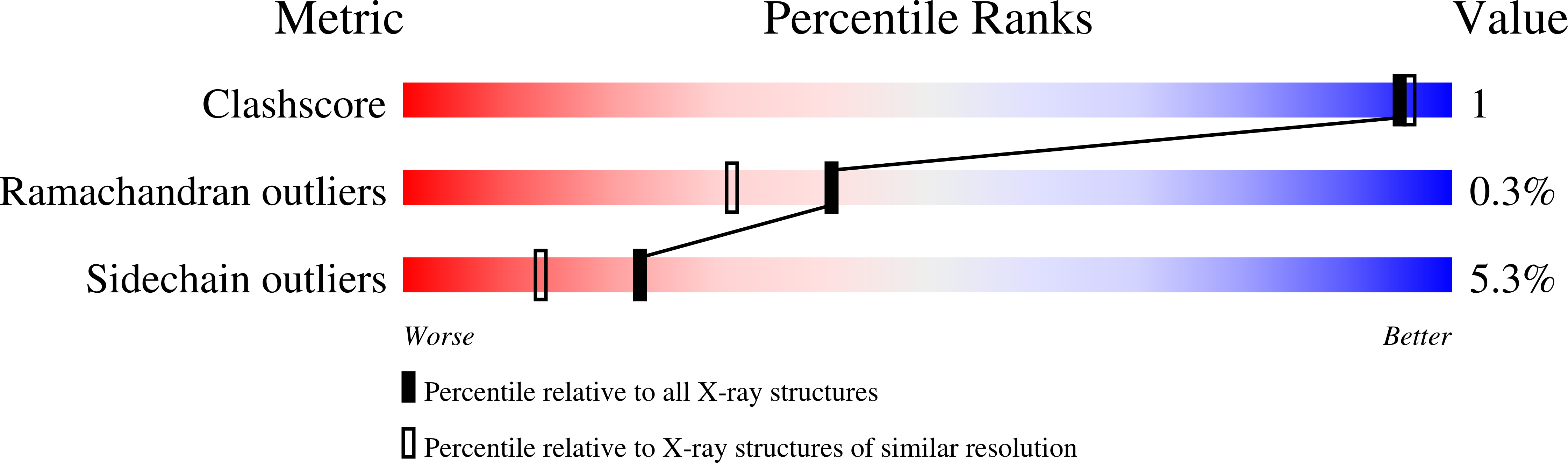
RCSB PDB - 8XIA: X-RAY ANALYSIS OF D-XYLOSE ISOMERASE AT 1.9 ANGSTROMS: NATIVE ENZYME IN COMPLEX WITH SUBSTRATE AND WITH A MECHANISM-DESIGNED INACTIVATOR
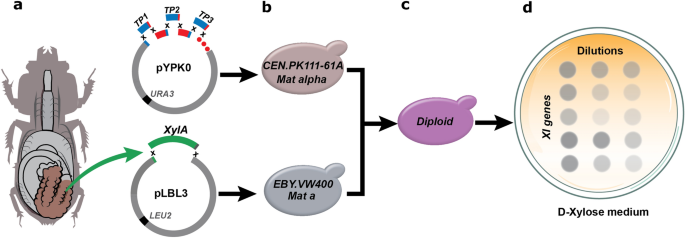
A novel d-xylose isomerase from the gut of the wood feeding beetle Odontotaenius disjunctus efficiently expressed in Saccharomyces cerevisiae | Scientific Reports

Probing the Roles of Active Site Residues in D-Xylose Isomerase (∗) - Journal of Biological Chemistry

Catalytic mechanism of xylose isomerase. Top: ring opening, middle:... | Download Scientific Diagram
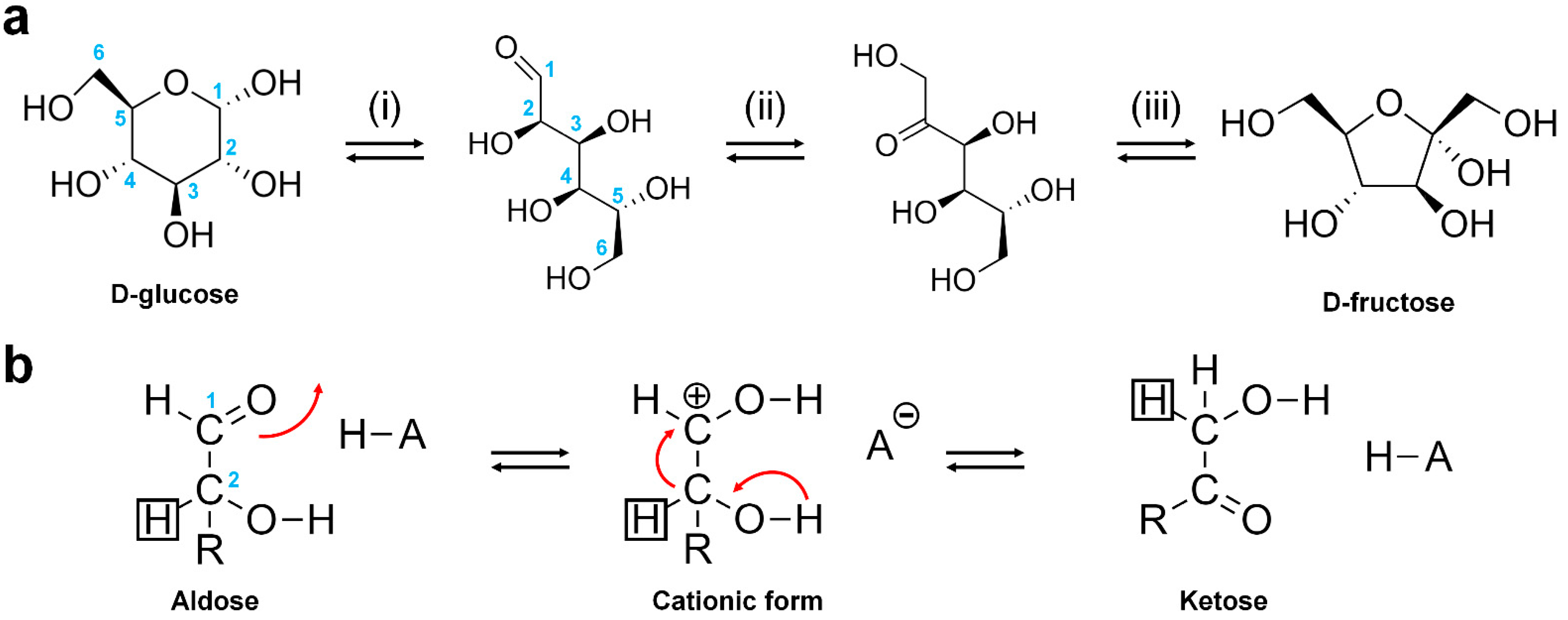
Applied Sciences | Free Full-Text | Glucose Isomerase: Functions, Structures, and Applications | HTML

Catalytic mechanism of xylose isomerase. Top: ring opening, middle:... | Download Scientific Diagram

Hydride transfer catalyzed by xylose isomerase: Mechanism and quantum effects - Garcia‐Viloca - 2003 - Journal of Computational Chemistry - Wiley Online Library

L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study: Structure

Figure 9 from Catalytic mechanism of xylose (glucose) isomerase from Clostridium thermosulfurogenes. Characterization of the structural gene and function of active site histidine. | Semantic Scholar

L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study: Structure

Xylose fermentation efficiency of industrial Saccharomyces cerevisiae yeast with separate or combined xylose reductase/xylitol dehydrogenase and xylose isomerase pathways | Biotechnology for Biofuels and Bioproducts | Full Text